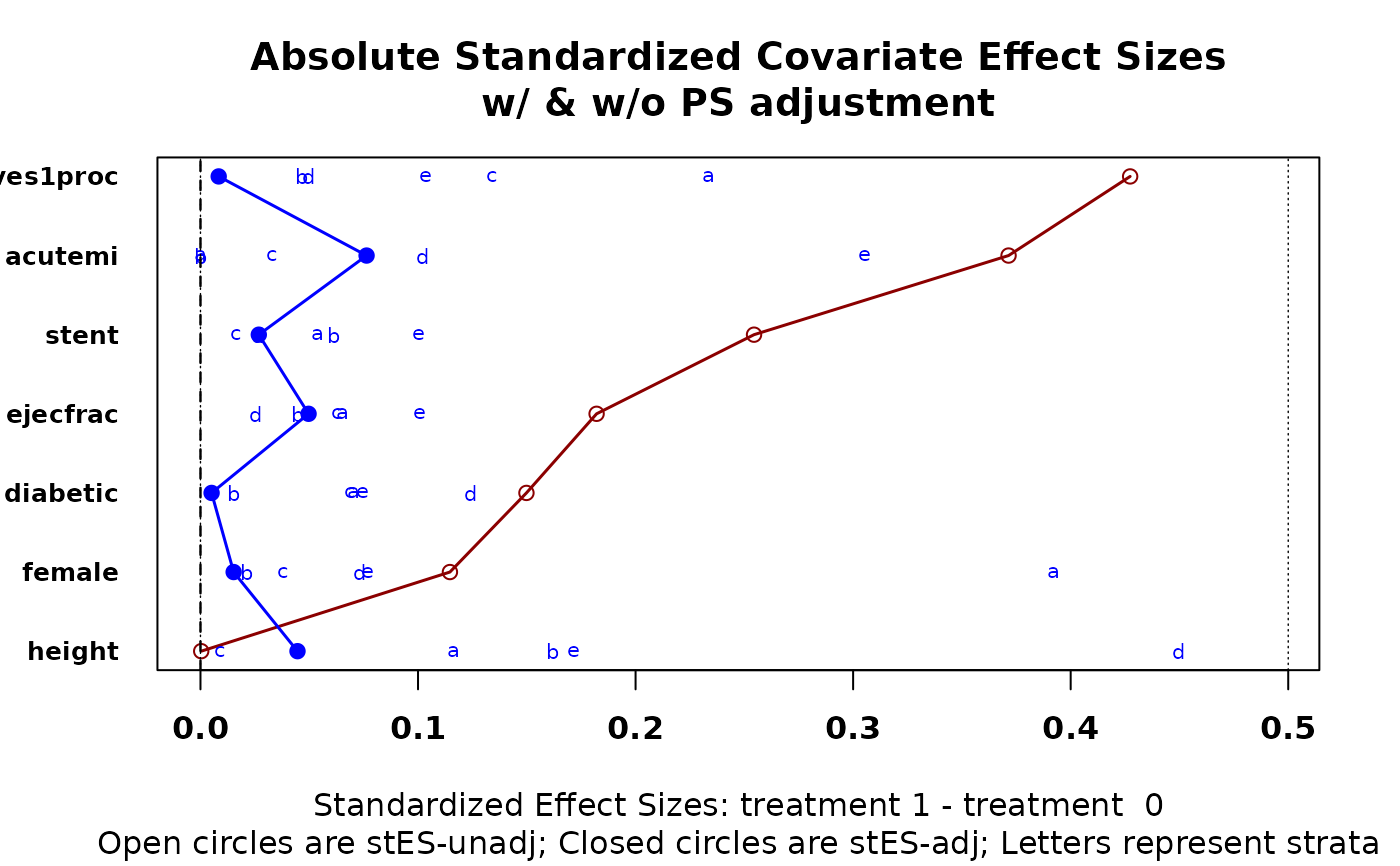
Provides a graphic that depicts covarite effect size differences between treatment groups both before and after stratification. Function will create stata internally if desired, and returns numerical output used to create graphic.
cv.bal.psa(
covariates,
treatment,
propensity,
strata = NULL,
int = NULL,
tree = FALSE,
minsize = 2,
universal.psd = TRUE,
trM = 0,
absolute.es = TRUE,
trt.value = NULL,
use.trt.var = FALSE,
verbose = FALSE,
xlim = NULL,
plot.strata = TRUE,
...
)Arguments
- covariates
Dataframe of covariates. Factors should be recoded using
cv.trans.psa- treatment
Binary vector or factor defining the two treatments
- propensity
Vector of same length as
treatmentcontaining estimated propensity scores.- strata
Either a vector of same length as
treatmentof predefined stratum number, or one integernused to assign rows tonstratapropensityscores, each of approximately the same number of cases. If relatively few unique propensity scores have been defined (as from a classification tree) then the logicaltreeshould be set equal toTRUE.- int
Either a number
mused to divide[0,1]intomequal length subintervals, or a vector containing cut points between 0 and 1 that define subintervals (perhaps as suggested by loess.psa). In either case the subintervals define strata, for which sizes can differ.- tree
Logical, default
FALSE. If there are few unique propensity scores, say from a recursively partitioned tree, then TRUE forces strata to be defined by the unique propensity scores.- minsize
Smallest allowable stratum-treatment size. If violated, rows in the stratum are removed. User may wish to redefine strata.
- universal.psd
Logical, default = TRUE. Forces standard deviations used to be unadjusted for stratification.
- trM
Numeric, default = 0; passed to
meanfor trimming purposes.- absolute.es
Logical, default TRUE. If TRUE, graphic depicts absolute values of all effect sizes. Note that the adjusted effect size plotted is the absolute value of weighted averages of the signed by-stratum effect size values when
absolute.esis TRUE.- trt.value
Character string; if desired allows the name of an active treatment to be given. Should be a level (value) of the
treatmentfactor (vector).- use.trt.var
Logical, default FALSE. If TRUE, uses just active treatment standard deviations for effect size, as per a suggestion of Rubin and Stuart (see reference below).
- verbose
Logical, default FALSE. Numerical output is returned invisibly.
- xlim
Binary vector passed to plot for overriding default choices. Default NULL.
- plot.strata
Logical, default TRUE. Adds effect size values for individual strata to graphic.
- ...
Other graphical parameters passed to
plot.
Value
Graphic plots covariate balance before and after stratication on
propensity scores. The default version (absolute.es = TRUE) plots the
absolute values of effect sizes for each stratum, though the overall
estimate is the weighted mean before taking the absolute values. Numerical
output consists of seven addressable objects. If verbose is FALSE
(default), output is not printed.
- original.strata
Matrix of strata-treatment counts as originally input.
- strata.used
Matrix of strata-treatment counts used in effectsize calculations after any
minsizereductions.- mean.diff.strata.wtd
Matrix of strata by covariate weighted (by strata size) average differences.
- mean.diff.unadj
Matrix of covariate effects sizes before stratification.
- effect.sizes
Matrix of effect sizes by covariate and statum.
- treatment.levels
Names of treatments.
- effects.strata.treatment
Matrix of standard deviations and stratum-treatment covariate means used to calculate the
effect.sizes.
Details
Effect sizes between treatments for each covariate are presented in one graphic, both before and after stratification.
References
``Matching Methods for Causal Inference: A review and a look forward." Forthcoming in Statistical Science.
See also
cv.bal.psa, loess.psa,
cstrata.psa, cv.trans.psa
Examples
data(lindner)
attach(lindner)
#> The following objects are masked from lindner (pos = 3):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
#> The following objects are masked from lindner (pos = 4):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
#> The following objects are masked from lindner (pos = 5):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
#> The following objects are masked from lindner (pos = 6):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.cv <- lindner[,4:10]
cv.bal.psa(lindner.cv, abcix, ps, strata = 5)
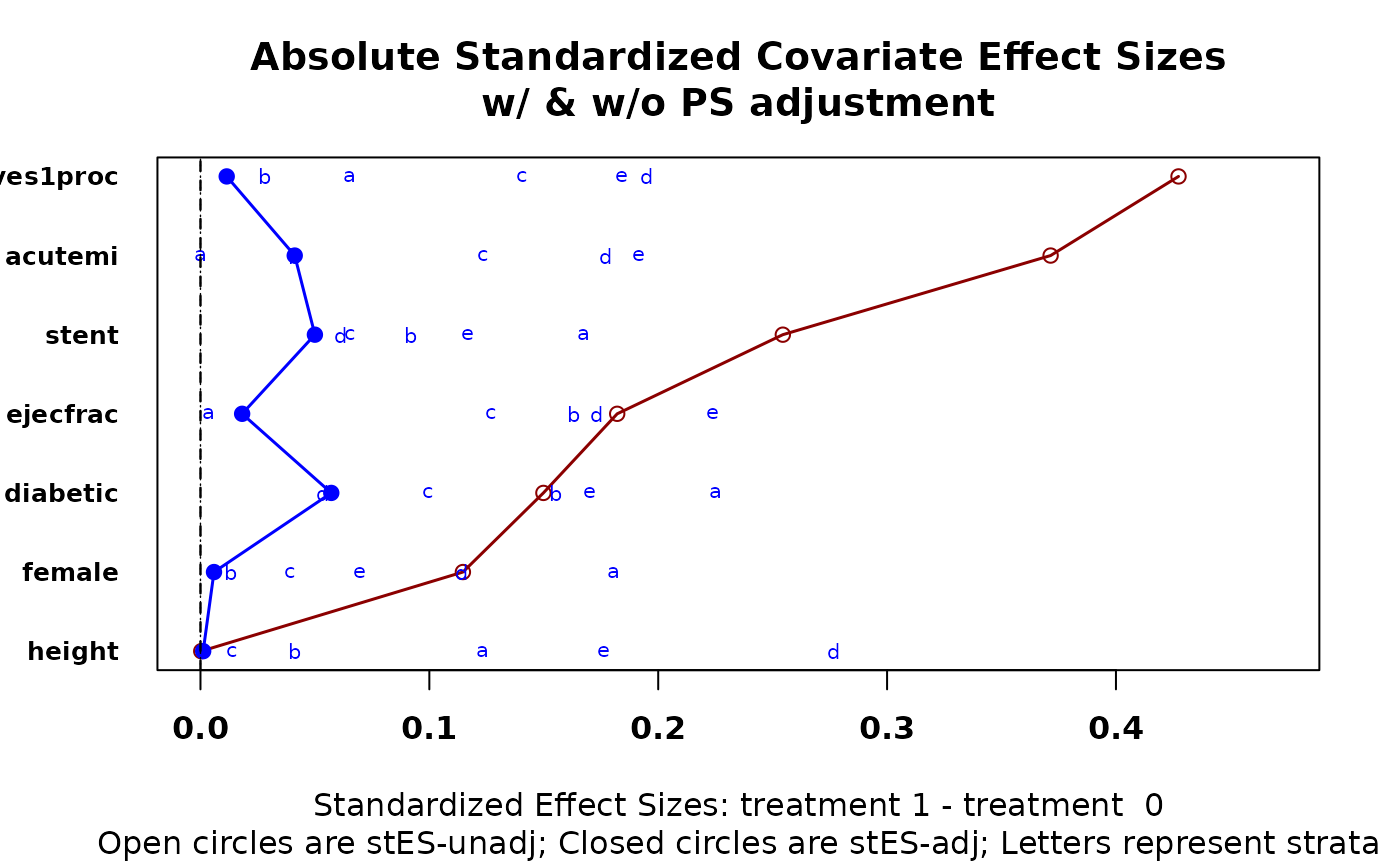 cv.bal.psa(lindner.cv, abcix, ps, strata = 10)
cv.bal.psa(lindner.cv, abcix, ps, strata = 10)
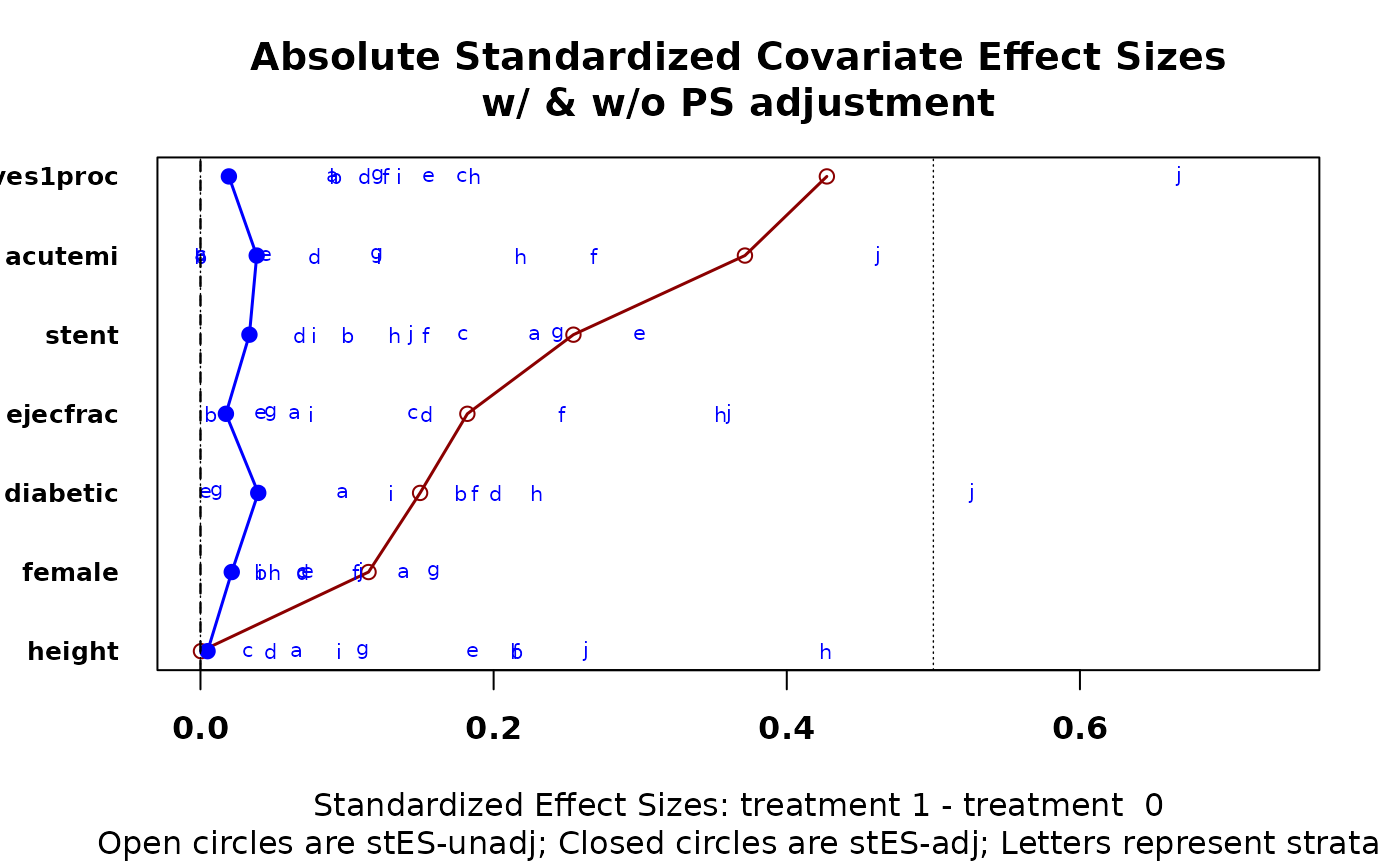 cv.bal.psa(lindner.cv, abcix, ps, int = c(.2, .5, .6, .75, .8))
cv.bal.psa(lindner.cv, abcix, ps, int = c(.2, .5, .6, .75, .8))