Displays a graphic that summarizes outcomes in a propensity score analysis, based on strata that have been defined in the first Phase of a propensity score analysis (PSA). The graphic displays contributions of individual strata to the overall effect, weighing contributions of individual strata according to the relative sizes of the respective strata. The overall effect is plotted as a heavy dashed diagonal line that runs parallel to the identity diagonal.
circ.psa(
response,
treatment = NULL,
strata = NULL,
summary = FALSE,
statistic = "mean",
trim = 0,
revc = FALSE,
confint = TRUE,
sw = 0.4,
ne = 0.5,
inc = 0.25,
pw = 0.4,
lab = TRUE,
labcex = 1,
xlab = NULL,
ylab = NULL,
main = NULL,
mai = c(1, 1.7, 1, 1.7)
)Arguments
- response
Either a numeric vector containing the response of interest in a propensity score analysis, or a three column array containing response, treatment and strata.
- treatment
Binary variable of same length as
response; generally 0 for 'control,' 1 for 'treatment'. A character vector with two labels or factor with two levels are also accepted.- strata
Generally integer variable; a vector of same length as
responseindicating the derived strata from estimated propensity scores. Generally 5 or 6 strata used, but function is effective for more strata. In the case when strata are defined via unique propensity scores (as from a tree), user may wish to define strata usingfactor.- summary
Logical (default
FALSE). IfTRUEthenresponsemust have rows corresponding to number of strata; the first two columns should contain treatment and control group sizes for each stratum, and the pair of columns should contain the appropriate summary statistics for each statum. For example, the four summary columns might have been generated by thestrata.summaryoutput ofloess.psa.- statistic
A scalar summary of the center of the response distribution. Seen next item below. Default = "mean". Note that to generate this statistic the full vector of responses must have been input, not summaries.
- trim
Allows for a trimmed mean as outcome measure, where trim is from 0 to .5 (.5 implying median).
- revc
Logical; if
TRUEthen X and Y axes are interchanged in plot.- confint
Logical; if
TRUEadds an approximate 95% confidence interval for the mean. The interval may not be realistic it thetrimargument exceeds zero.- sw
Numerical argument (default = 0.4); extends axes on lower ends, effectively moving circles to lower left.
- ne
Numerical argument (default = 0.5); extends axes on upper ends, effectively moving circles to upper right.
- inc
Numerical argument (default = 0.35); controls circle sizes, but relative circle sizes are controlled via
pw. In general one wants circle areas to appear subjectively to be sized in accordance with strata sizes.- pw
numerical argument (default = 0.4); controls relative circle sizes.
pwdenotes power or exponent for radius of circle.- lab
Logical (default TRUE); labels circles with stratum labels.
- labcex
numerical argument (default = 1); controls the size of the circle labels.
- xlab
Label for horizontal axis, by default taken from
treatment.- ylab
Label for vertical axis, by default taken from
treatment.- main
Main label for graph.
- mai
margin parameters.
Value
Generate a Propensity Assessment Plot, as well as numerical data for
- summary.strata
An array with rows corresponding to strata and four columns; these show counts for control and treatment groups, as well as (possibly trimmed) mean response values for control and treatment.
- wtd.Mn.(Name1)
Weighted mean of response for (Name1) group. Name taken from
treatment.- wtd.Mn.(Name2)
Weighted mean of response for (Name2) group. Name taken from
treatment.- ATE
Average Treatment Effect.
- se.wtd
Weighted standard error for
ATE- approx.t
Ratio of the average treatement effect and a standard error based on weighting of stratum variances.
- df
Estimate of degree of freedom; response vector length minus twice number of strata.
- CI.95
Approximate 95% confidence interval for overall effect size.
Details
A circle is plotted for each stratum, centered on the means for the
treatment and control groups (for the X and Y axes)
respectively. The sizes of the circles correspond to the relative sizes of
the strata. A diagonal line (lower left to upper right) shows the identity,
X=Y, so that circles on, say, the lower side of this line show that
the corresponding X mean is larger than the Y mean for that
stratum, and vice-versa. Parallel projections are made from the centers of
the strata-cum-circles to difference scores that are plotted on a line
segment on the lower-left corner of the graphic; the average difference,
which corresponds to the average treatment effect (ATE) for the overall
treatment effect, is plotted as a heavy (dark blue) dashed line parallel to
the identity diagonal. Rug plots are shown on the upper and right margins of
the graphic, for the X and Y marginal distributions. A 95%
confidence interval for the overall effect is plotted to the left of the
distribution of the stratum difference scores, centered on the ATE. Trimmed
means can replace the conventional mean for both the ATE and the marginal
distributions (however, the confidence interval calculations are likely to
become less trustworthy as larger values of the trim argument are used).
See also
Examples
##Random data with effect size 0
response <- rnorm(1000)
treatment <- sample(c(0,1), 1000, replace = TRUE)
strata <- sample(1:6, 1000, replace = TRUE)
circ.psa(response, treatment, strata)
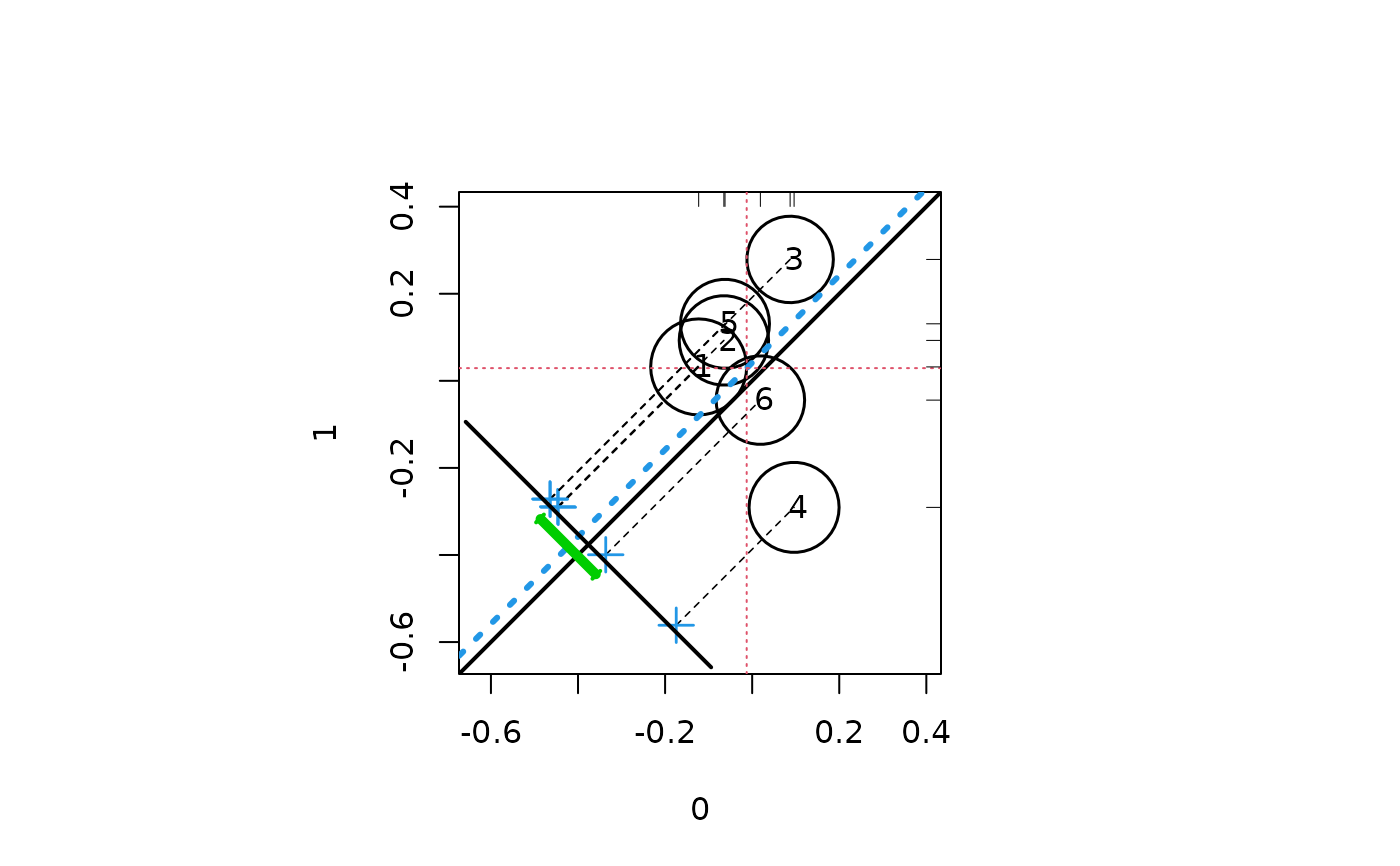 #> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 92 104 -0.12287194 0.03195197
#> 2 82 82 -0.06495047 0.09283901
#> 3 62 89 0.08714321 0.27879215
#> 4 78 89 0.09625740 -0.29066839
#> 5 73 89 -0.06241920 0.13097107
#> 6 72 88 0.01883228 -0.04430640
#>
#> $wtd.Mn.0
#> [1] -0.01259991
#>
#> $wtd.Mn.1
#> [1] 0.02917246
#>
#> $ATE
#> [1] 0.04177238
#>
#> $se.wtd
#> [1] 0.0646297
#>
#> $approx.t
#> [1] 0.6463341
#>
#> $df
#> [1] 988
#>
#> $CI.95
#> [1] -0.08505488 0.16859964
#>
##Random data with effect size -.2
response <- c(rnorm(500, 0, 12), rnorm(500, 6, 12))
treatment <- c(rep(0, 500), rep(1,500))
strata <- sample(1:5, 1000, replace = TRUE)
aaa <- cbind(response, treatment, strata)
circ.psa(aaa)
#> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 92 104 -0.12287194 0.03195197
#> 2 82 82 -0.06495047 0.09283901
#> 3 62 89 0.08714321 0.27879215
#> 4 78 89 0.09625740 -0.29066839
#> 5 73 89 -0.06241920 0.13097107
#> 6 72 88 0.01883228 -0.04430640
#>
#> $wtd.Mn.0
#> [1] -0.01259991
#>
#> $wtd.Mn.1
#> [1] 0.02917246
#>
#> $ATE
#> [1] 0.04177238
#>
#> $se.wtd
#> [1] 0.0646297
#>
#> $approx.t
#> [1] 0.6463341
#>
#> $df
#> [1] 988
#>
#> $CI.95
#> [1] -0.08505488 0.16859964
#>
##Random data with effect size -.2
response <- c(rnorm(500, 0, 12), rnorm(500, 6, 12))
treatment <- c(rep(0, 500), rep(1,500))
strata <- sample(1:5, 1000, replace = TRUE)
aaa <- cbind(response, treatment, strata)
circ.psa(aaa)
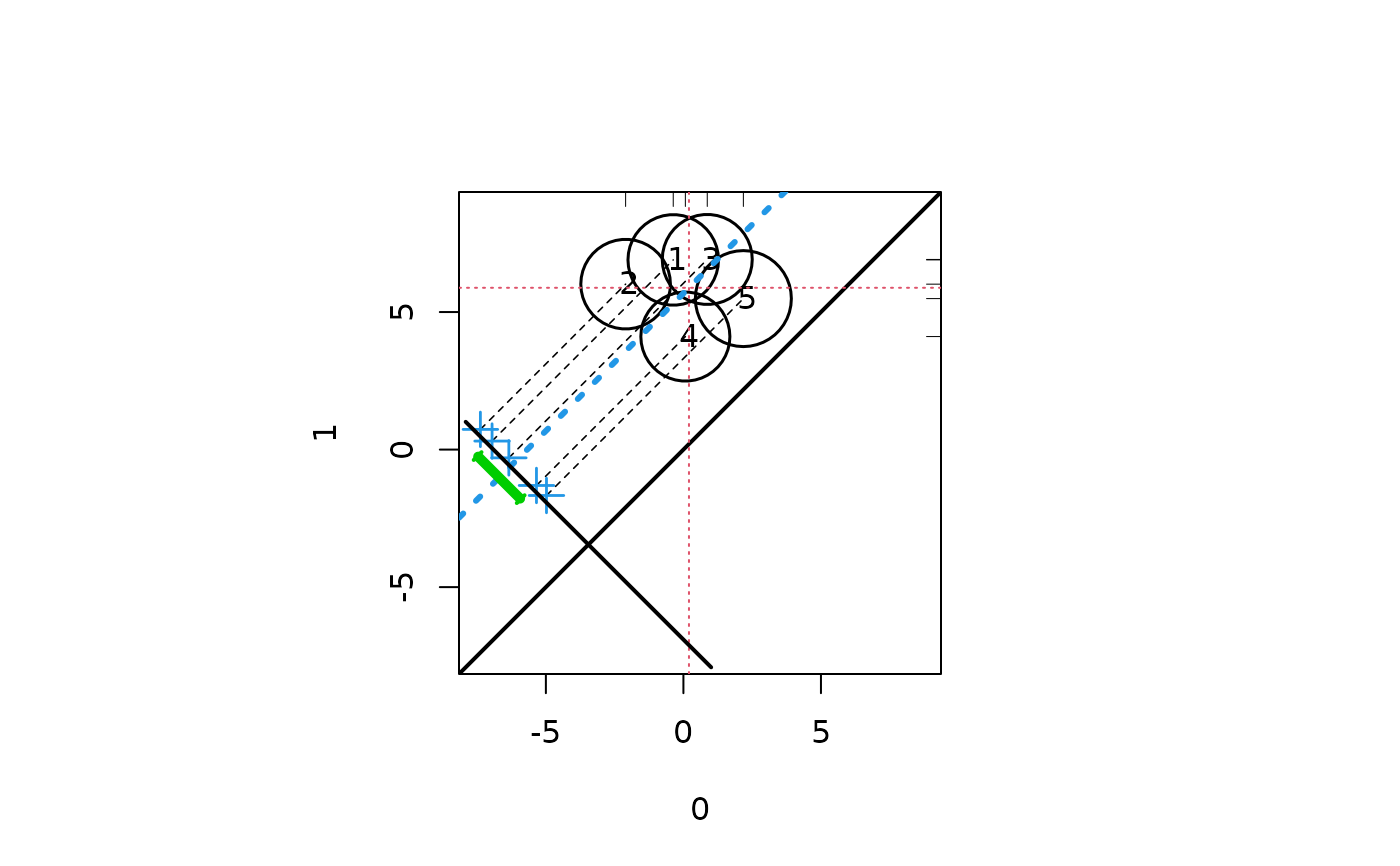 #> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 102 95 -0.36992232 6.895803
#> 2 88 104 -2.10107829 6.014000
#> 3 100 94 0.86670728 6.912477
#> 4 84 105 0.07079183 4.108854
#> 5 126 102 2.17934013 5.488770
#>
#> $wtd.Mn.0
#> [1] 0.2021287
#>
#> $wtd.Mn.1
#> [1] 5.882194
#>
#> $ATE
#> [1] 5.680066
#>
#> $se.wtd
#> [1] 0.777143
#>
#> $approx.t
#> [1] 7.308907
#>
#> $df
#> [1] 990
#>
#> $CI.95
#> [1] 4.155029 7.205102
#>
##Random data with effect size -2
response <- c(rt(100,3) * 2 + 20, rt(100,12) * 2 + 18)
treatment <- rep(c("A","B"), each = 100)
strata <- sample(c("X","Y","Z","U","V"), 200, replace = TRUE)
circ.psa(response, treatment, strata)
#> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 102 95 -0.36992232 6.895803
#> 2 88 104 -2.10107829 6.014000
#> 3 100 94 0.86670728 6.912477
#> 4 84 105 0.07079183 4.108854
#> 5 126 102 2.17934013 5.488770
#>
#> $wtd.Mn.0
#> [1] 0.2021287
#>
#> $wtd.Mn.1
#> [1] 5.882194
#>
#> $ATE
#> [1] 5.680066
#>
#> $se.wtd
#> [1] 0.777143
#>
#> $approx.t
#> [1] 7.308907
#>
#> $df
#> [1] 990
#>
#> $CI.95
#> [1] 4.155029 7.205102
#>
##Random data with effect size -2
response <- c(rt(100,3) * 2 + 20, rt(100,12) * 2 + 18)
treatment <- rep(c("A","B"), each = 100)
strata <- sample(c("X","Y","Z","U","V"), 200, replace = TRUE)
circ.psa(response, treatment, strata)
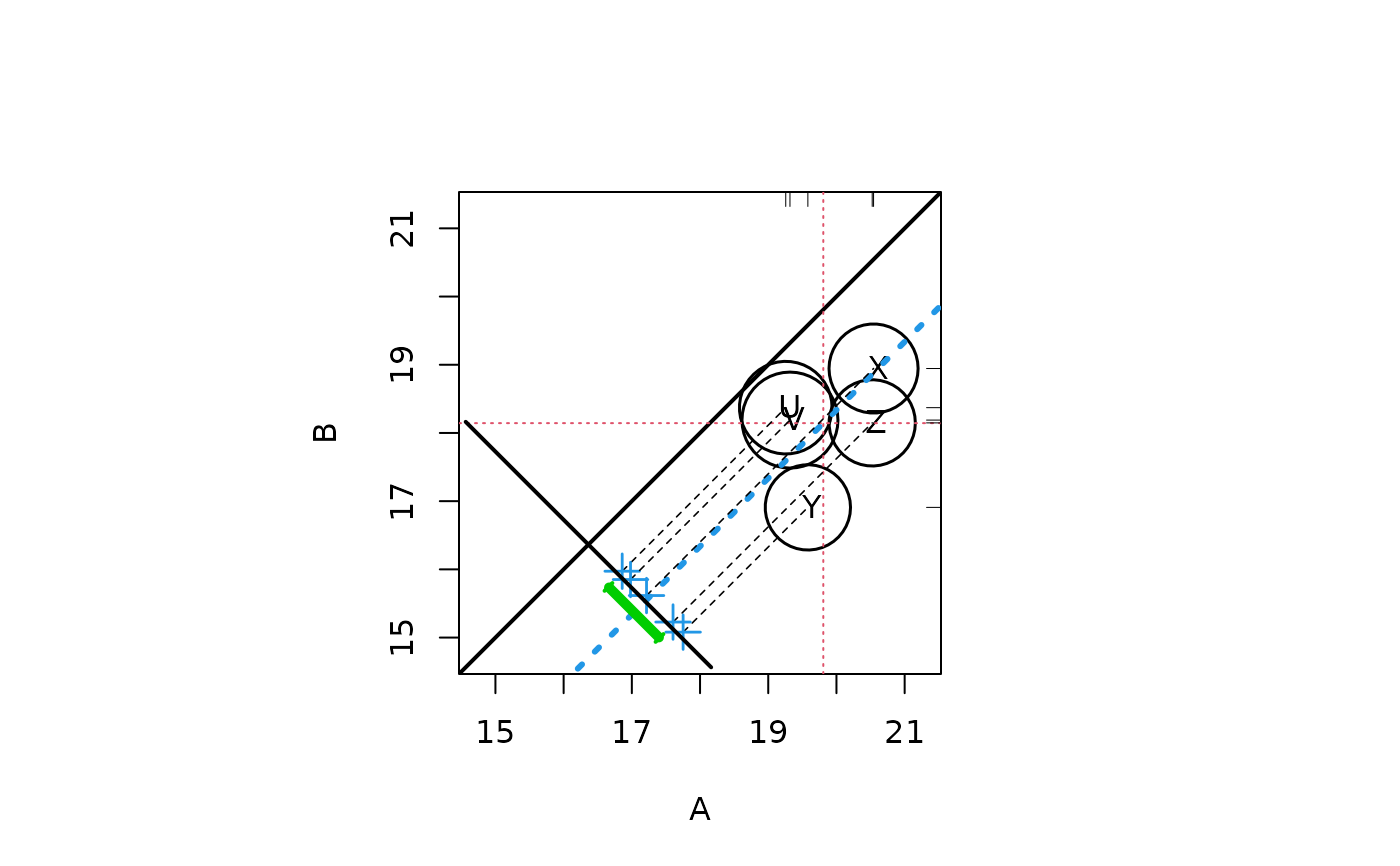 #> $summary.strata
#> n.A n.B means.A means.B
#> U 21 22 19.25658 18.37083
#> V 21 26 19.31841 18.18866
#> X 21 18 20.54407 18.94484
#> Y 19 16 19.58090 16.90878
#> Z 18 18 20.52424 18.14787
#>
#> $wtd.Mn.A
#> [1] 19.80711
#>
#> $wtd.Mn.B
#> [1] 18.14396
#>
#> $ATE
#> [1] -1.663146
#>
#> $se.wtd
#> [1] 0.3709823
#>
#> $approx.t
#> [1] -4.483088
#>
#> $df
#> [1] 190
#>
#> $CI.95
#> [1] -2.3949194 -0.9313734
#>
##Tree derived strata
library(rpart)
data(lindner)
attach(lindner)
#> The following objects are masked from lindner (pos = 3):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
#> The following objects are masked from lindner (pos = 4):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
lindner.rpart <- rpart(abcix ~ stent + height + female + diabetic +
acutemi + ejecfrac + ves1proc, data = lindner, method = "class")
lindner.tree<-factor(lindner.rpart$where, labels = 1:6)
circ.psa(log(cardbill), abcix, lindner.tree)
#> $summary.strata
#> n.A n.B means.A means.B
#> U 21 22 19.25658 18.37083
#> V 21 26 19.31841 18.18866
#> X 21 18 20.54407 18.94484
#> Y 19 16 19.58090 16.90878
#> Z 18 18 20.52424 18.14787
#>
#> $wtd.Mn.A
#> [1] 19.80711
#>
#> $wtd.Mn.B
#> [1] 18.14396
#>
#> $ATE
#> [1] -1.663146
#>
#> $se.wtd
#> [1] 0.3709823
#>
#> $approx.t
#> [1] -4.483088
#>
#> $df
#> [1] 190
#>
#> $CI.95
#> [1] -2.3949194 -0.9313734
#>
##Tree derived strata
library(rpart)
data(lindner)
attach(lindner)
#> The following objects are masked from lindner (pos = 3):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
#> The following objects are masked from lindner (pos = 4):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
lindner.rpart <- rpart(abcix ~ stent + height + female + diabetic +
acutemi + ejecfrac + ves1proc, data = lindner, method = "class")
lindner.tree<-factor(lindner.rpart$where, labels = 1:6)
circ.psa(log(cardbill), abcix, lindner.tree)
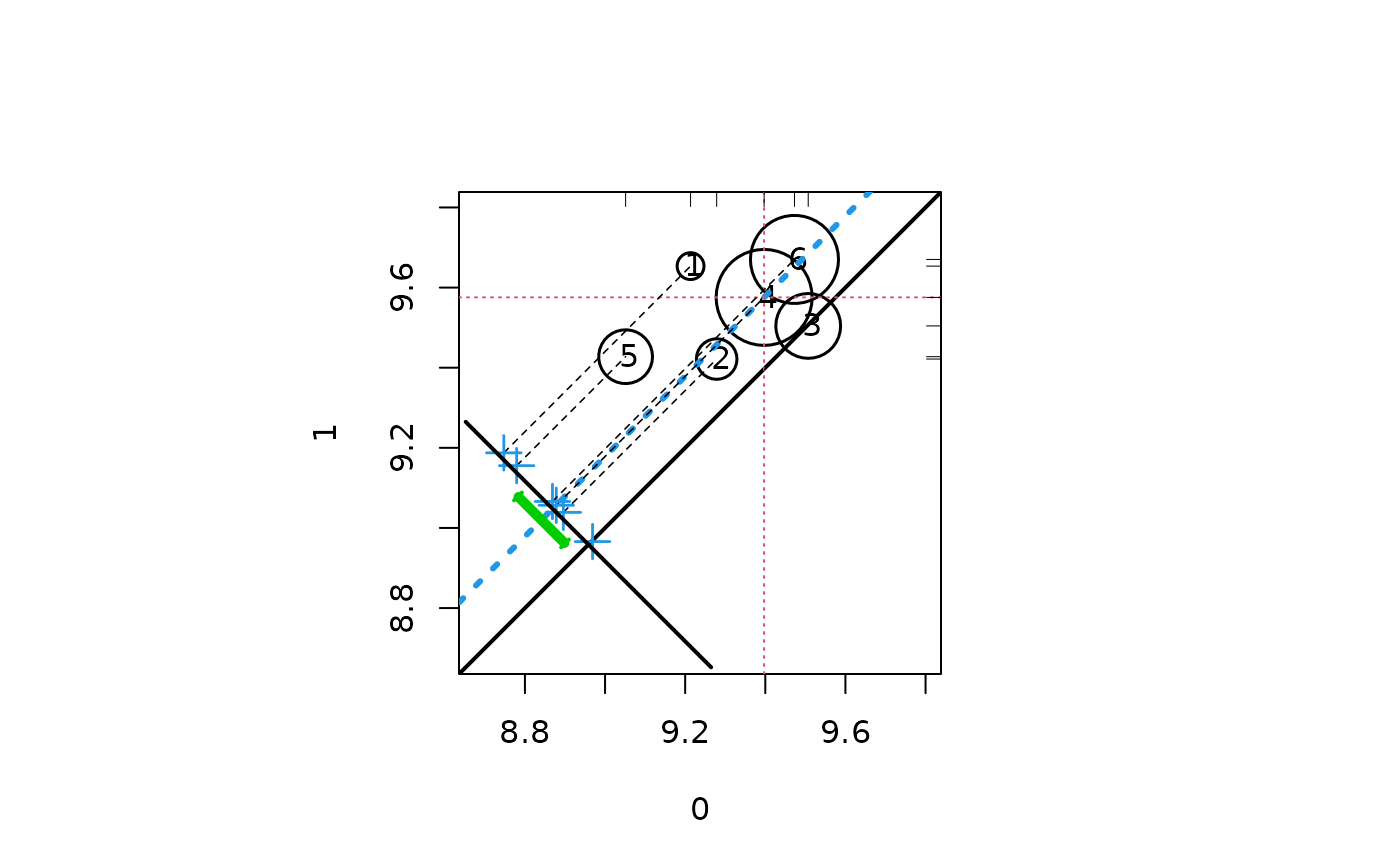 #> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 14 2 9.213456 9.653571
#> 2 29 16 9.278627 9.421610
#> 3 58 87 9.507137 9.504301
#> 4 131 256 9.397094 9.575469
#> 5 12 79 9.051331 9.427481
#> 6 54 258 9.472841 9.670249
#>
#> $wtd.Mn.0
#> [1] 9.396949
#>
#> $wtd.Mn.1
#> [1] 9.57558
#>
#> $ATE
#> [1] 0.1786314
#>
#> $se.wtd
#> [1] 0.05838647
#>
#> $approx.t
#> [1] 3.059466
#>
#> $df
#> [1] 984
#>
#> $CI.95
#> [1] 0.06405511 0.29320771
#>
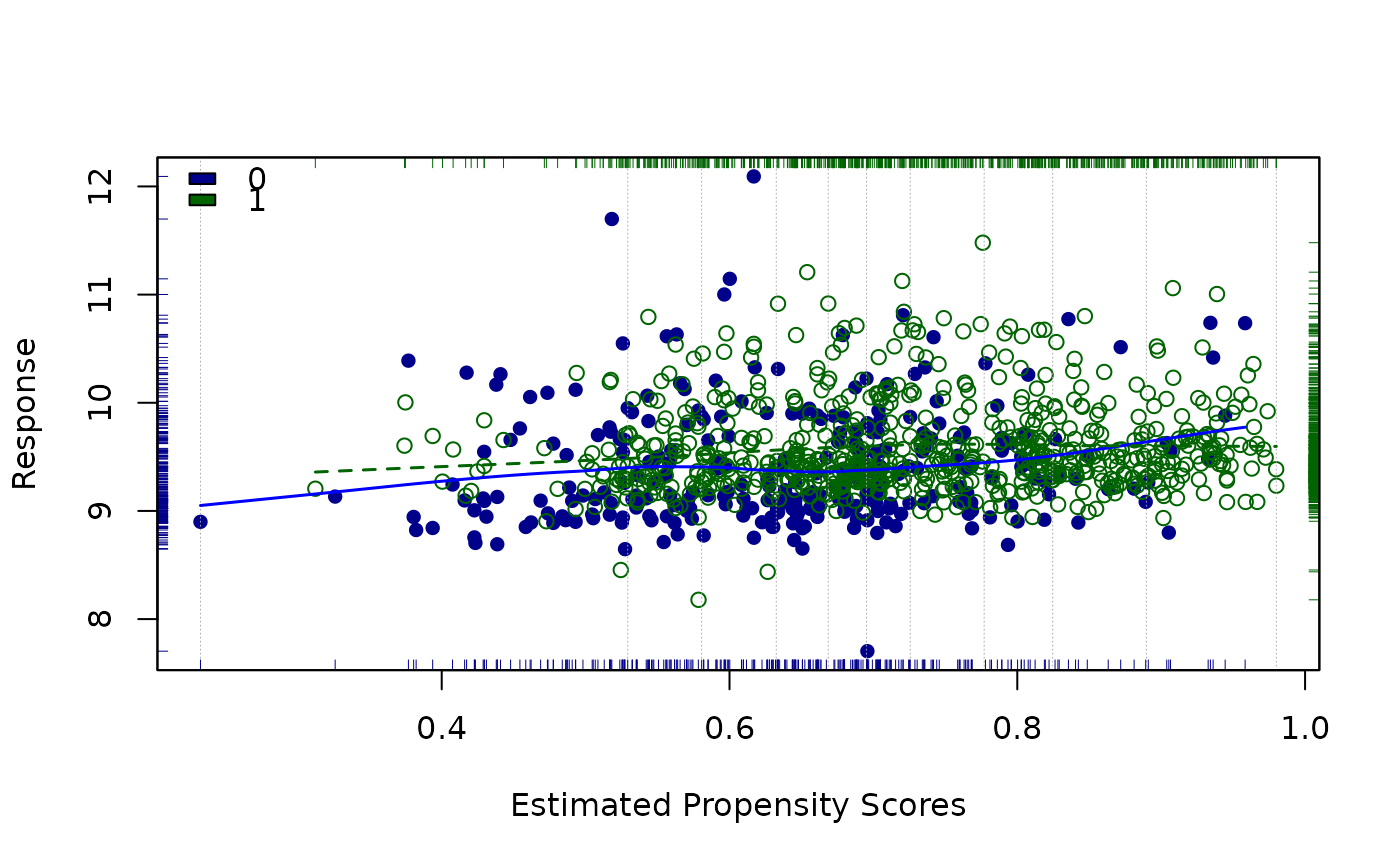
##Loess derived strata
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.loess<-loess.psa(log(cardbill), abcix, ps)
#> $summary.strata
#> n.0 n.1 means.0 means.1
#> 1 14 2 9.213456 9.653571
#> 2 29 16 9.278627 9.421610
#> 3 58 87 9.507137 9.504301
#> 4 131 256 9.397094 9.575469
#> 5 12 79 9.051331 9.427481
#> 6 54 258 9.472841 9.670249
#>
#> $wtd.Mn.0
#> [1] 9.396949
#>
#> $wtd.Mn.1
#> [1] 9.57558
#>
#> $ATE
#> [1] 0.1786314
#>
#> $se.wtd
#> [1] 0.05838647
#>
#> $approx.t
#> [1] 3.059466
#>
#> $df
#> [1] 984
#>
#> $CI.95
#> [1] 0.06405511 0.29320771
#>
##Loess derived strata
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.loess<-loess.psa(log(cardbill), abcix, ps)
 circ.psa(lindner.loess$summary.strata[, 1:4], summary = TRUE,
inc = .1, labcex = .7)
circ.psa(lindner.loess$summary.strata[, 1:4], summary = TRUE,
inc = .1, labcex = .7)
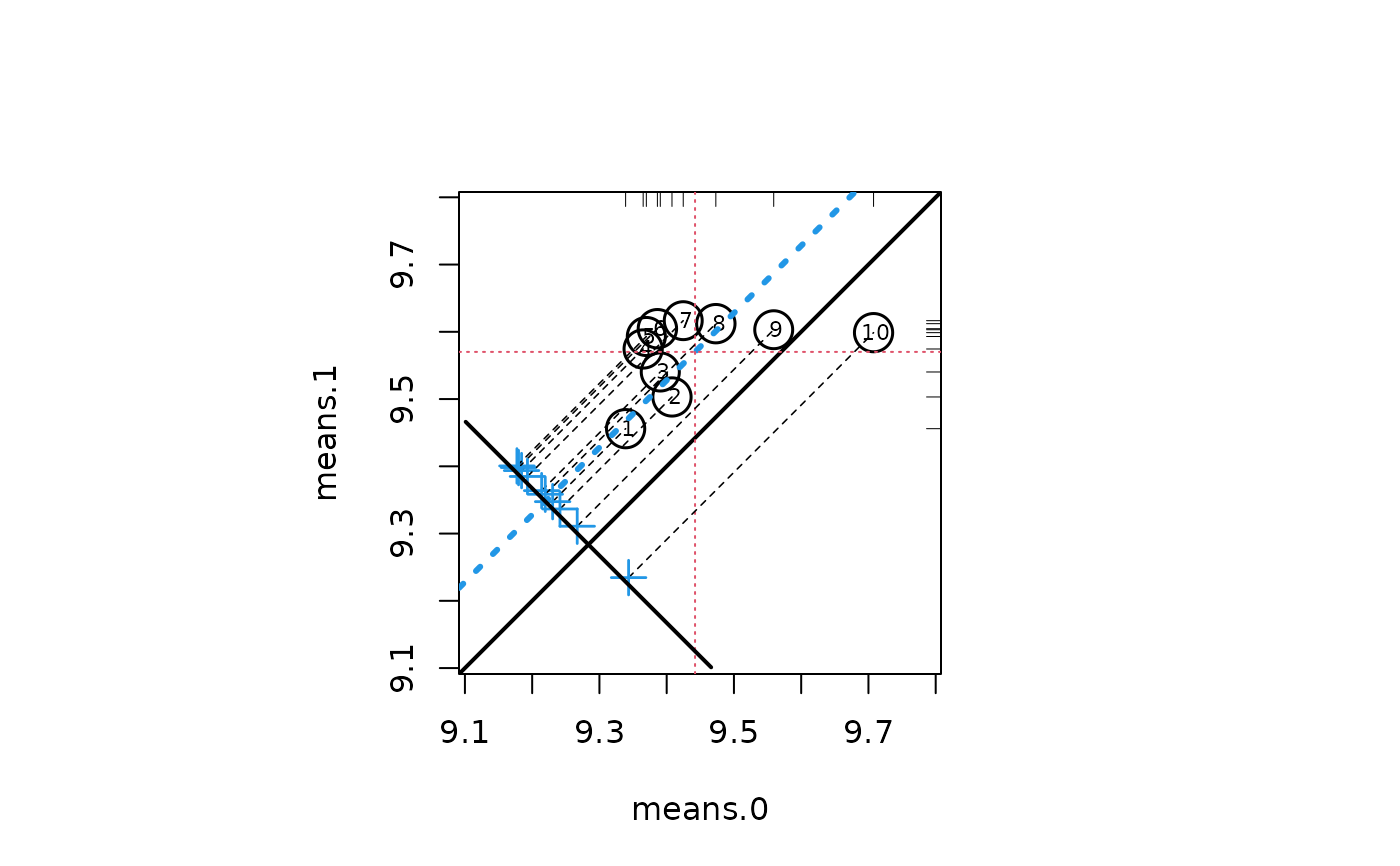 #> $summary.strata
#> counts.0 counts.1 means.0 means.1
#> 1 58 42 9.338939 9.455965
#> 2 37 63 9.407877 9.503125
#> 3 35 64 9.390499 9.540272
#> 4 40 60 9.365029 9.574458
#> 5 29 70 9.369748 9.593021
#> 6 36 65 9.386188 9.604613
#> 7 24 74 9.424611 9.616556
#> 8 19 82 9.473078 9.612104
#> 9 11 87 9.559145 9.603100
#> 10 9 91 9.707606 9.598751
#>
#> $wtd.Mn.means.0
#> [1] 9.442172
#>
#> $wtd.Mn.means.1
#> [1] 9.570121
#>
#> $ATE
#> [1] 0.1279487
#>
#> $se.wtd
#> NULL
#>
#> $approx.t
#> numeric(0)
#>
#> $df
#> [1] 976
#>
#> $CI.95
#> [1] NA NA
#>
#> $summary.strata
#> counts.0 counts.1 means.0 means.1
#> 1 58 42 9.338939 9.455965
#> 2 37 63 9.407877 9.503125
#> 3 35 64 9.390499 9.540272
#> 4 40 60 9.365029 9.574458
#> 5 29 70 9.369748 9.593021
#> 6 36 65 9.386188 9.604613
#> 7 24 74 9.424611 9.616556
#> 8 19 82 9.473078 9.612104
#> 9 11 87 9.559145 9.603100
#> 10 9 91 9.707606 9.598751
#>
#> $wtd.Mn.means.0
#> [1] 9.442172
#>
#> $wtd.Mn.means.1
#> [1] 9.570121
#>
#> $ATE
#> [1] 0.1279487
#>
#> $se.wtd
#> NULL
#>
#> $approx.t
#> numeric(0)
#>
#> $df
#> [1] 976
#>
#> $CI.95
#> [1] NA NA
#>