Compare balance graphically of a categorical covariate as part of a PSA
Source:R/cat.psa.R
cat.psa.RdGiven predefined strata and two level treatment for a categorical covariate
from a propensity score analysis, cat.psa draws pairs of side by side
barplots corresponding to control and treatment for each stratum.
cat.psa(
categorical,
treatment = NULL,
strata = NULL,
catnames = NULL,
catcol = "terrain.colors",
width = 0.25,
barlab = c("A", "B"),
barnames = NULL,
rtmar = 1.5,
balance = FALSE,
B = 1000,
tbl = TRUE,
cex.leg = 1,
...
)Arguments
- categorical
Vector or N X 3 dataframe or matrix. If a vector, then represents a categorical covariate that is being balanced within strata in a PSA. If
categoricalhas three columns, then the second and third are assumed to be thetreatmentandstratarespectively. Missing values are not allowed. May be factor or numeric.- treatment
Binary vector or factor of same length as
continuousrepresenting the two treatments.- strata
A vector or factor of same length as
continuousindicating the derived strata from estimated propensity scores. Strata are ordered lexicographically in plot.- catnames
List of names in order of the categories; used in the plot legend. Default is
1:n.- catcol
List of colors used for the categories, default is
terrain.colors.- width
Controls width of bars, default = 0.25.
- barlab
Binary list of single
treatmentcharacter labels for the bars, default isc("A", "B"). These are defined in a legend bybarnames.- barnames
Binary list of treatment names used in the legend; by default names are taken from
treatment.- rtmar
Numeric. Governs size of right margin allocated for legend. Default = 1.5
- balance
Logical. If
TRUEa call is made to functionsbal.cs.psaandbal.cws.psa. The former provides a reference histogram and ad hoc balance statistic, the second provides bootstrapped p-values for the two-way table formed in each statum. Default isFALSE.- B
Numeric; passed to
bal.cs.psagoverning size of reference histogram generated. Default is 100.- tbl
Logical; if
TRUE, then a matrix of the proportions used in the creation of the bargraph is returned.- cex.leg
Numeric; value of
cex(governing font size) passed to legend. Default = 1.- ...
Other graphical parameters passed to plot.
Value
If tbl is TRUE, then a matrix is returned containing
the proportions of each category, and in each treatment level and stratum
that were used to draw the bargraph.
Details
Pairs of bars are graphed side by side so that comparisons may be made
within each stratum and across strata. If balance is TRUE,
then the histogram represents an ad hoc balance measure of the given strata
as compared to randomly generated strata. The p-values provided on the
bargraph are bootstrapped in a standard fashion via randomly generated
treatment divisions within given strata. For continuous covariates use
box.psa.
See also
bal.cs.psa, bal.cws.psa, box.psa
Examples
categorical<-sample(1:7,1000,replace=TRUE)
treatment<-sample(c(0,1),1000,replace=TRUE)
strata<-sample(5,1000,replace=TRUE)
cat.psa(categorical,treatment,strata)
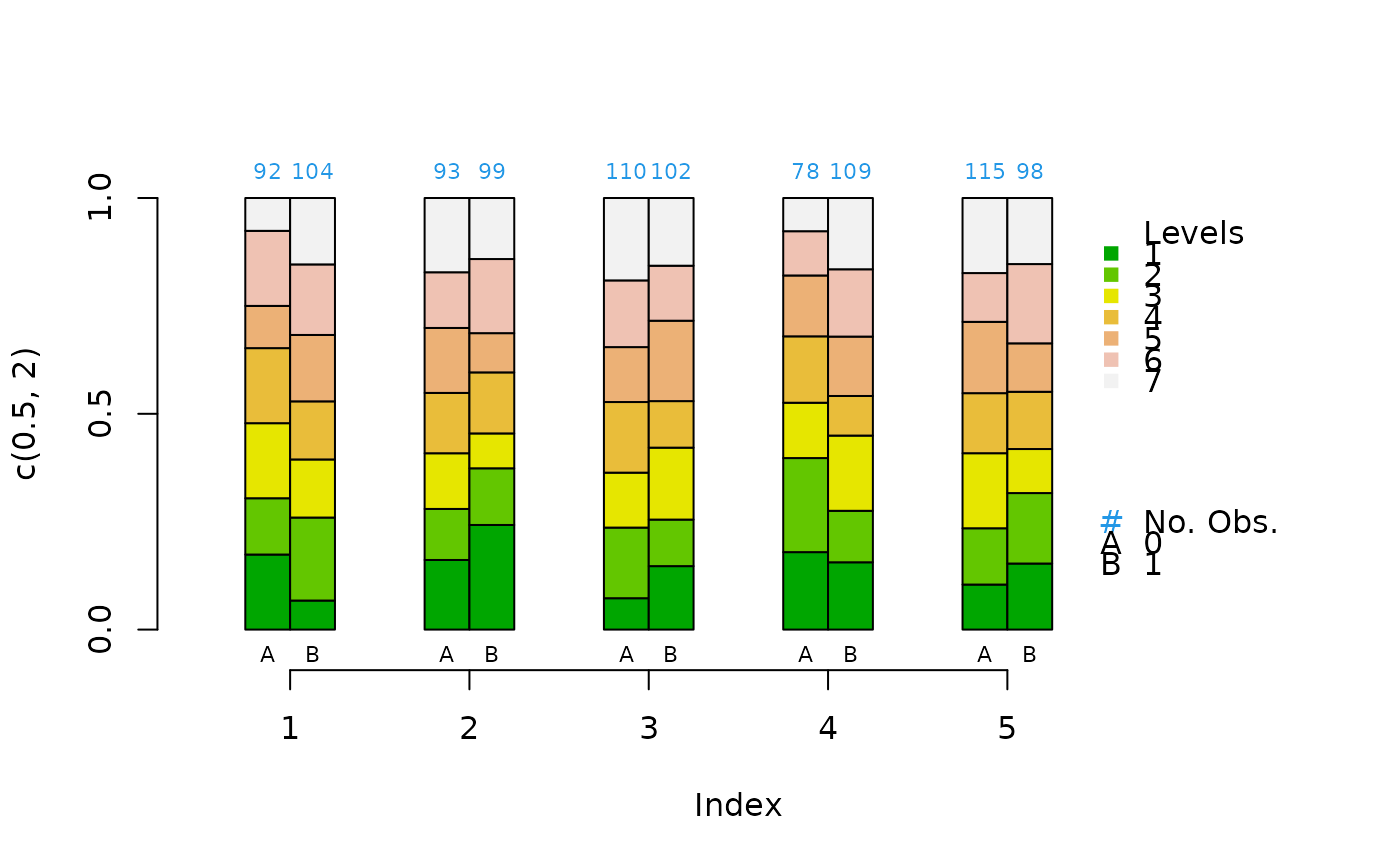 #> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5
#> 1 0.174 0.067 0.161 0.242 0.073 0.147 0.179 0.156 0.104 0.153
#> 2 0.130 0.192 0.118 0.131 0.164 0.108 0.218 0.119 0.130 0.163
#> 3 0.174 0.135 0.129 0.081 0.127 0.167 0.128 0.174 0.174 0.102
#> 4 0.174 0.135 0.140 0.141 0.164 0.108 0.154 0.092 0.139 0.133
#> 5 0.098 0.154 0.151 0.091 0.127 0.186 0.141 0.138 0.165 0.112
#> 6 0.174 0.163 0.129 0.172 0.155 0.127 0.103 0.156 0.113 0.184
#> 7 0.076 0.154 0.172 0.141 0.191 0.157 0.077 0.165 0.174 0.153
#>
data(lindner)
attach(lindner)
#> The following objects are masked from lindner (pos = 3):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.s5 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
include.lowest = TRUE, labels = FALSE))
cat.psa(stent, abcix, lindner.s5, xlab = "stent")
#> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5
#> 1 0.174 0.067 0.161 0.242 0.073 0.147 0.179 0.156 0.104 0.153
#> 2 0.130 0.192 0.118 0.131 0.164 0.108 0.218 0.119 0.130 0.163
#> 3 0.174 0.135 0.129 0.081 0.127 0.167 0.128 0.174 0.174 0.102
#> 4 0.174 0.135 0.140 0.141 0.164 0.108 0.154 0.092 0.139 0.133
#> 5 0.098 0.154 0.151 0.091 0.127 0.186 0.141 0.138 0.165 0.112
#> 6 0.174 0.163 0.129 0.172 0.155 0.127 0.103 0.156 0.113 0.184
#> 7 0.076 0.154 0.172 0.141 0.191 0.157 0.077 0.165 0.174 0.153
#>
data(lindner)
attach(lindner)
#> The following objects are masked from lindner (pos = 3):
#>
#> abcix, acutemi, cardbill, diabetic, ejecfrac, female, height,
#> lifepres, stent, ves1proc
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.s5 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
include.lowest = TRUE, labels = FALSE))
cat.psa(stent, abcix, lindner.s5, xlab = "stent")
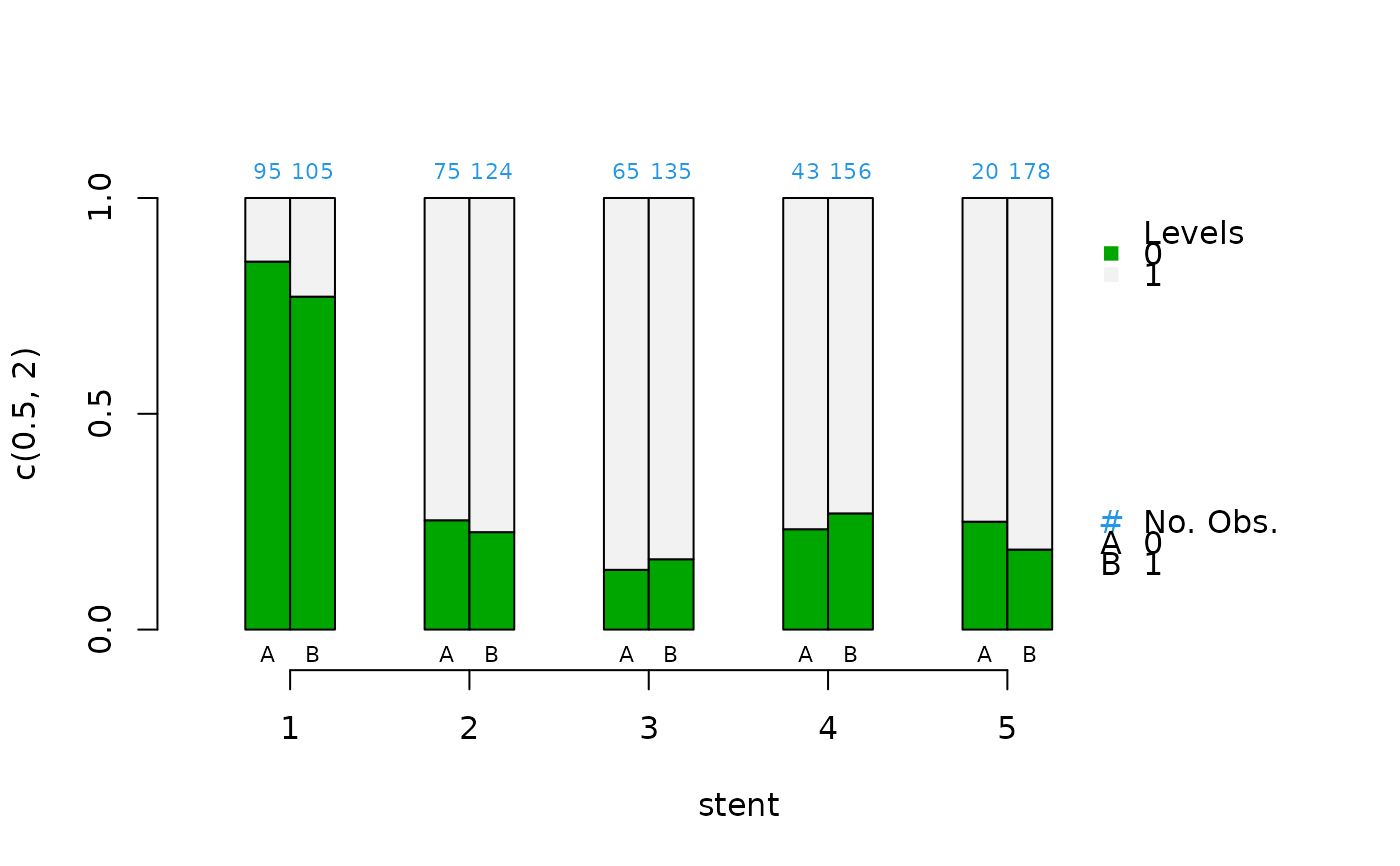 #> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5
#> 0 0.853 0.771 0.253 0.226 0.138 0.163 0.233 0.269 0.25 0.185
#> 1 0.147 0.229 0.747 0.774 0.862 0.837 0.767 0.731 0.75 0.815
#>
lindner.s10 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/10)),
include.lowest = TRUE, labels = FALSE))
cat.psa(ves1proc,abcix, lindner.s10, balance = TRUE, xlab = "ves1proc")
#> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5
#> 0 0.853 0.771 0.253 0.226 0.138 0.163 0.233 0.269 0.25 0.185
#> 1 0.147 0.229 0.747 0.774 0.862 0.837 0.767 0.731 0.75 0.815
#>
lindner.s10 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/10)),
include.lowest = TRUE, labels = FALSE))
cat.psa(ves1proc,abcix, lindner.s10, balance = TRUE, xlab = "ves1proc")
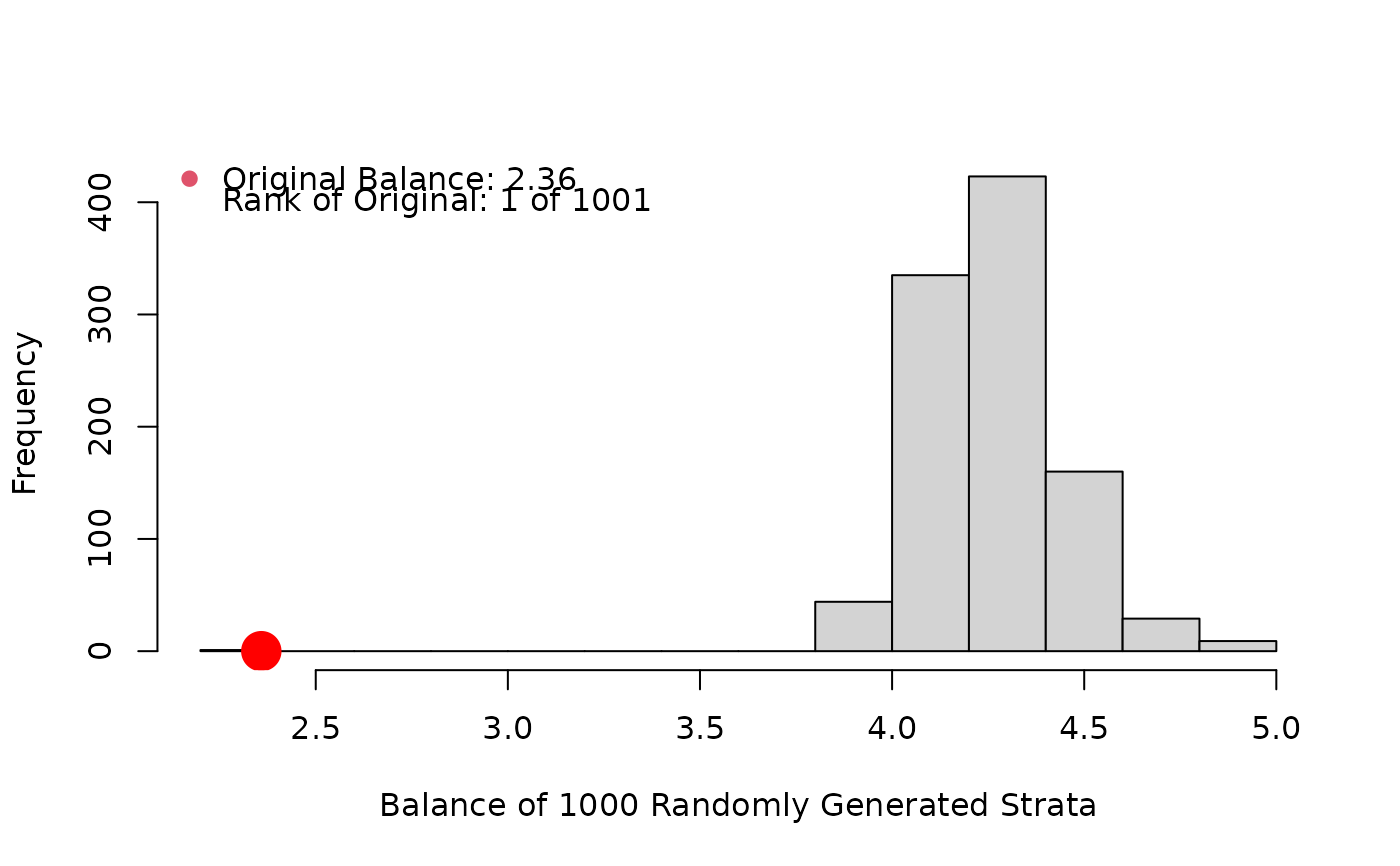 #> Histogram of Random Strata Balance. Press <enter> for next chart...
#> Histogram of Random Strata Balance. Press <enter> for next chart...
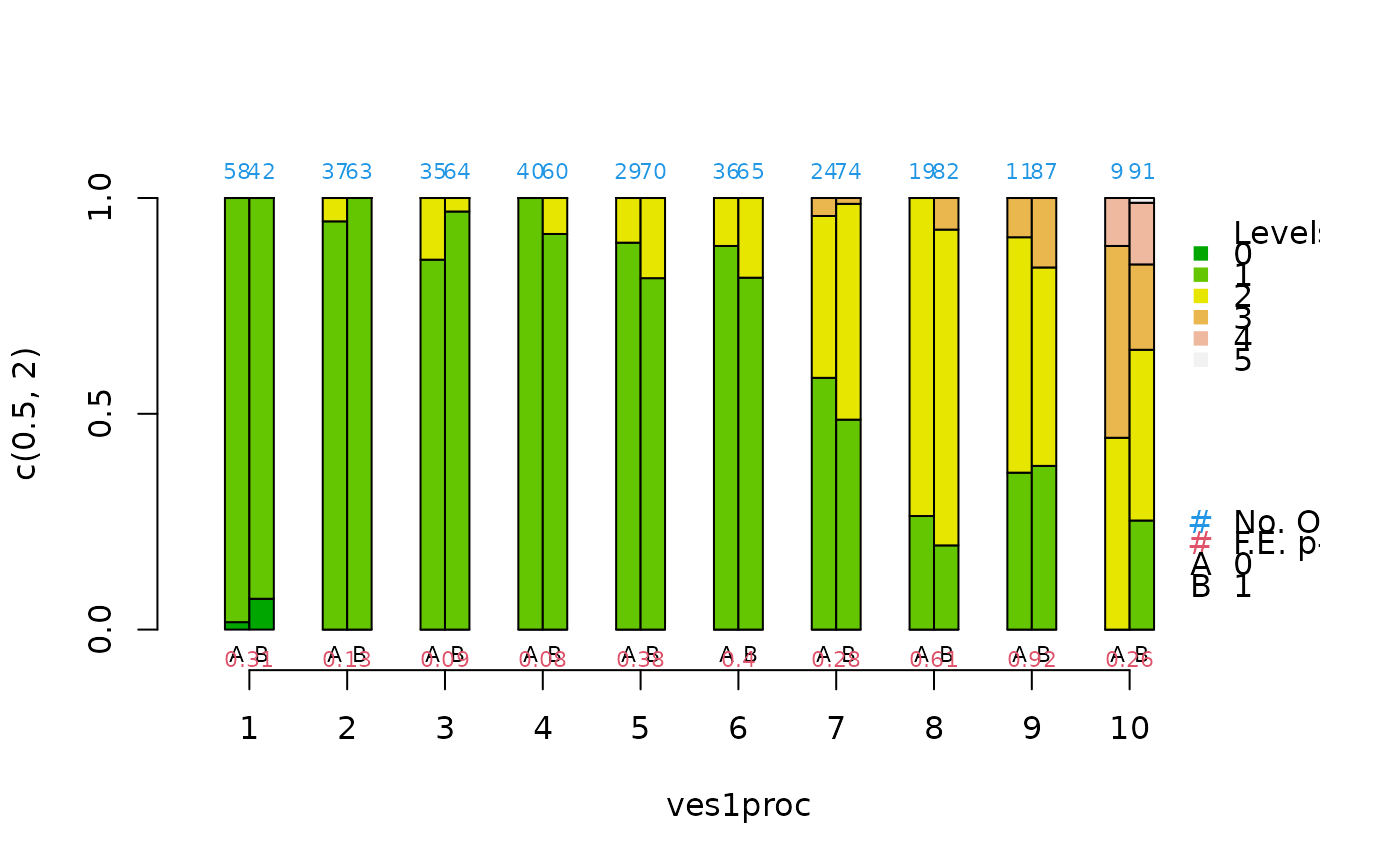 #> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6 0:7
#> 0 0.017 0.071 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 1 0.983 0.929 0.946 1 0.857 0.969 1 0.917 0.897 0.814 0.889 0.815 0.583
#> 2 0.000 0.000 0.054 0 0.143 0.031 0 0.083 0.103 0.186 0.111 0.185 0.375
#> 3 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.042
#> 4 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 5 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 1:7 0:8 1:8 0:9 1:9 0:10 1:10
#> 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000
#> 1 0.486 0.263 0.195 0.364 0.379 0.000 0.253
#> 2 0.500 0.737 0.732 0.545 0.460 0.444 0.396
#> 3 0.014 0.000 0.073 0.091 0.161 0.444 0.198
#> 4 0.000 0.000 0.000 0.000 0.000 0.111 0.143
#> 5 0.000 0.000 0.000 0.000 0.000 0.000 0.011
#>
#Using a rpart tree for strata
library(rpart)
lindner.rpart<-rpart(abcix ~ stent + height + female + diabetic +
acutemi + ejecfrac + ves1proc, data=lindner, method="class")
lindner.tree<-factor(lindner.rpart$where, labels = 1:6)
cat.psa(stent, abcix, lindner.tree, xlab = "stent")
#> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6 0:7
#> 0 0.017 0.071 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 1 0.983 0.929 0.946 1 0.857 0.969 1 0.917 0.897 0.814 0.889 0.815 0.583
#> 2 0.000 0.000 0.054 0 0.143 0.031 0 0.083 0.103 0.186 0.111 0.185 0.375
#> 3 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.042
#> 4 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 5 0.000 0.000 0.000 0 0.000 0.000 0 0.000 0.000 0.000 0.000 0.000 0.000
#> 1:7 0:8 1:8 0:9 1:9 0:10 1:10
#> 0 0.000 0.000 0.000 0.000 0.000 0.000 0.000
#> 1 0.486 0.263 0.195 0.364 0.379 0.000 0.253
#> 2 0.500 0.737 0.732 0.545 0.460 0.444 0.396
#> 3 0.014 0.000 0.073 0.091 0.161 0.444 0.198
#> 4 0.000 0.000 0.000 0.000 0.000 0.111 0.143
#> 5 0.000 0.000 0.000 0.000 0.000 0.000 0.011
#>
#Using a rpart tree for strata
library(rpart)
lindner.rpart<-rpart(abcix ~ stent + height + female + diabetic +
acutemi + ejecfrac + ves1proc, data=lindner, method="class")
lindner.tree<-factor(lindner.rpart$where, labels = 1:6)
cat.psa(stent, abcix, lindner.tree, xlab = "stent")
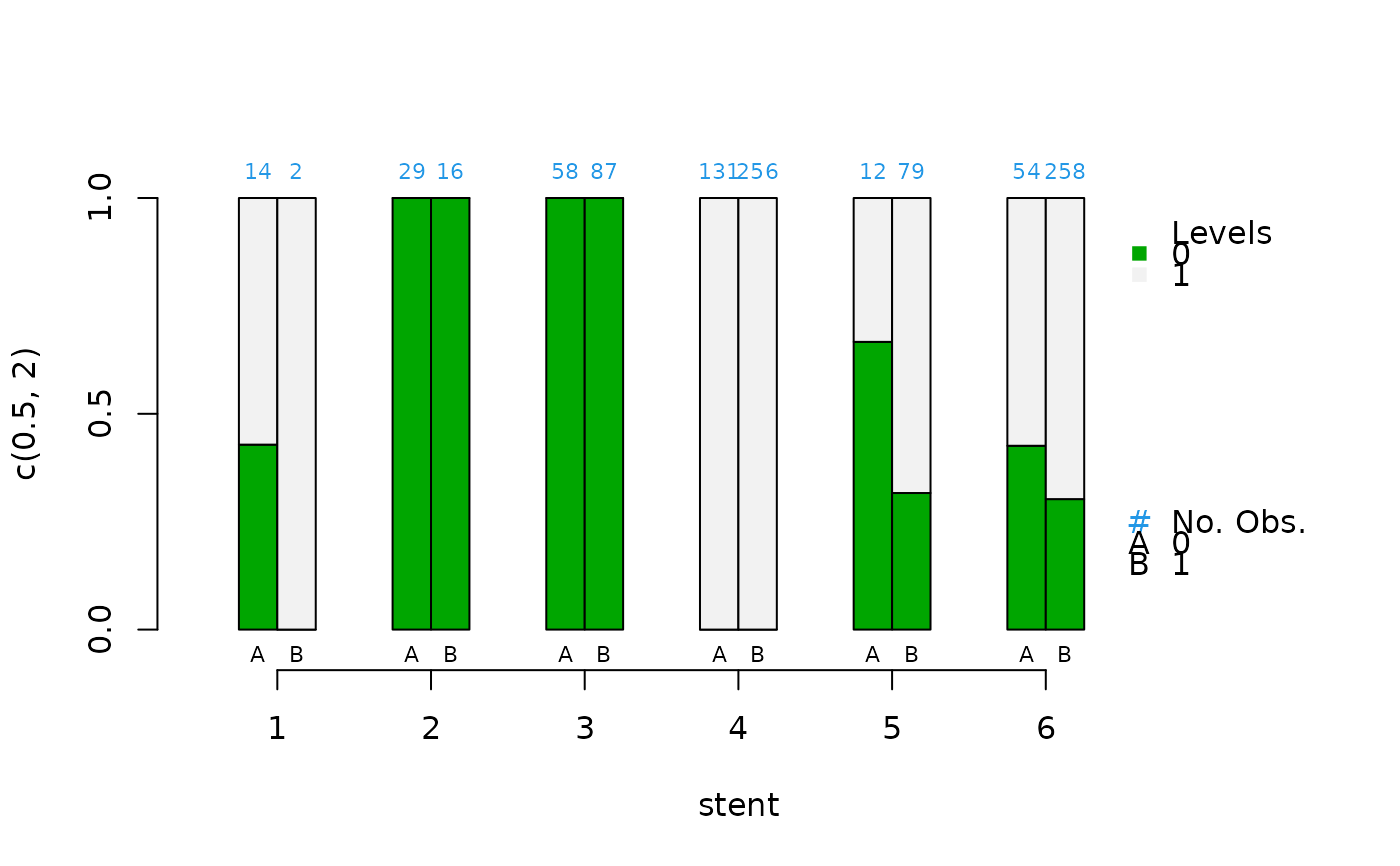 #> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6
#> 0 0.429 0 1 1 1 1 0 0 0.667 0.316 0.426 0.302
#> 1 0.571 1 0 0 0 0 1 1 0.333 0.684 0.574 0.698
#>
cat.psa(ves1proc, abcix, lindner.tree, xlab = "ves1proc")
#> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6
#> 0 0.429 0 1 1 1 1 0 0 0.667 0.316 0.426 0.302
#> 1 0.571 1 0 0 0 0 1 1 0.333 0.684 0.574 0.698
#>
cat.psa(ves1proc, abcix, lindner.tree, xlab = "ves1proc")
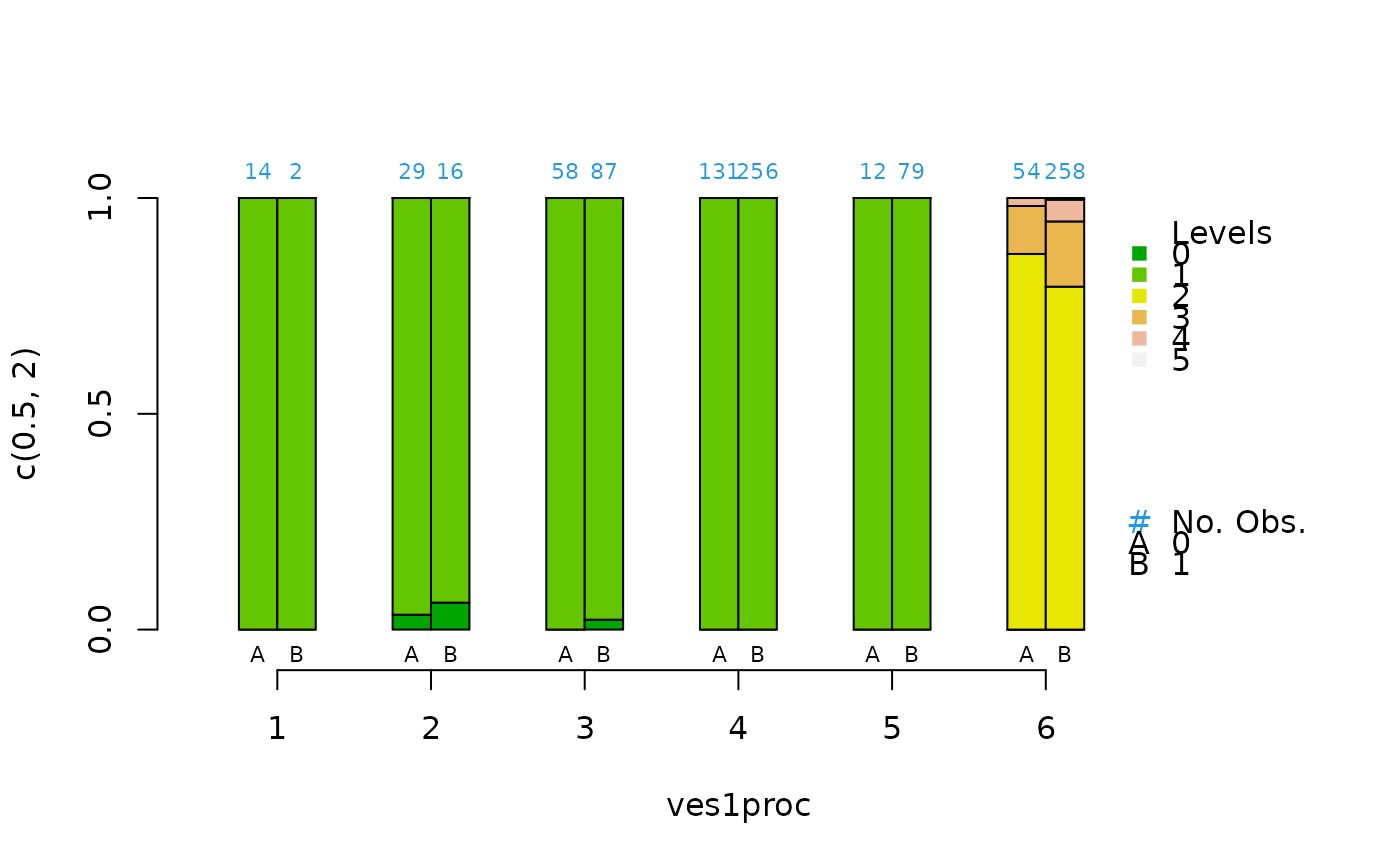 #> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6
#> 0 0 0 0.034 0.062 0 0.023 0 0 0 0 0.000 0.000
#> 1 1 1 0.966 0.938 1 0.977 1 1 1 1 0.000 0.000
#> 2 0 0 0.000 0.000 0 0.000 0 0 0 0 0.870 0.795
#> 3 0 0 0.000 0.000 0 0.000 0 0 0 0 0.111 0.151
#> 4 0 0 0.000 0.000 0 0.000 0 0 0 0 0.019 0.050
#> 5 0 0 0.000 0.000 0 0.000 0 0 0 0 0.000 0.004
#>
#> $`treatment:stratum.proportions`
#> 0:1 1:1 0:2 1:2 0:3 1:3 0:4 1:4 0:5 1:5 0:6 1:6
#> 0 0 0 0.034 0.062 0 0.023 0 0 0 0 0.000 0.000
#> 1 1 1 0.966 0.938 1 0.977 1 1 1 1 0.000 0.000
#> 2 0 0 0.000 0.000 0 0.000 0 0 0 0 0.870 0.795
#> 3 0 0 0.000 0.000 0 0.000 0 0 0 0 0.111 0.151
#> 4 0 0 0.000 0.000 0 0.000 0 0 0 0 0.019 0.050
#> 5 0 0 0.000 0.000 0 0.000 0 0 0 0 0.000 0.004
#>